sandbox/ghigo/src/test-stokes/poiseuille45.c
Poiseuille flow in a periodic channel inclined at 45 degrees
We solve here the viscous Stokes flow driven by gravity in an inclined periodic channel.
#include "../myembed.h"
#if CENTERED == 1
#include "../mycentered2.h"
#else
#include "../mycentered.h"
#endif // CENTERED
#include "view.h"Exact solution
We define here the exact solution for the velocity, evaluated at the center of each cell. Note that this expression is valid for the coordinate system along the direction of the channel of width H=0.5\, \cos(\frac{\pi}{4}).
static double exact (double y)
{
return sqrt(2)/2.*(sq ((0.5/sqrt (2.))/2.) - sq (y)); // ||G|| = sqrt(2), mu = 1, H = 0.5/sqrt(2)
}We also define the shape of the domain. We shift the position of the walls by EPS along the y-x direction to avoid pathological cases for the definition of the embedded boundaries where the embedded boundary exactly passes through two diagonaly opposed corners of the cell.
#define EPS (1.e-14)
void p_shape (scalar c, face vector f)
{
vertex scalar phi[];
foreach_vertex()
phi[] = difference (
union (
y - x - EPS,
x - y - 0.5 + EPS),
y - x - 0.5 + EPS);
fractions (phi, c, f);
fractions_cleanup (c, f,
smin = 1.e-14, cmin = 1.e-14);
}
#if TREEWhen using TREE, we try to increase the accuracy of the restriction operation in pathological cases by defining the gradient of u at the center of the cell.
void u_embed_gradient_x (Point point, scalar s, coord * g)
{
double xy = (y - x) >= 0. ? (y - x) - 0.25 : (y - x) + 0.75;
xy /= sqrt (2.);
g->x = cos (pi/4.)*(-sqrt (2.)/2.)*(2.*xy)*(-1./sqrt (2.));
g->y = cos (pi/4.)*(-sqrt (2.)/2.)*(2.*xy)*( 1./sqrt (2.));
}
void u_embed_gradient_y (Point point, scalar s, coord * g)
{
double xy = (y - x) >= 0. ? (y - x) - 0.25 : (y - x) + 0.75;
xy /= sqrt (2.);
g->x = sin (pi/4.)*(-sqrt (2.)/2.)*(2.*xy)*(-1./sqrt (2.));
g->y = sin (pi/4.)*(-sqrt (2.)/2.)*(2.*xy)*( 1./sqrt (2.));
}
#endif // TREESetup
We need a field for viscosity so that the embedded boundary metric can be taken into account.
face vector muv[];We also define a reference velocity field.
scalar un[];
int lvl;
int main()
{The domain is 1\times 1 and periodic.
We set the maximum timestep.
DT = 1.e-1;We set the tolerance of the Poisson solver.
stokes = true;
TOLERANCE = 1.e-7;
TOLERANCE_MU = 1.e-7;
for (lvl = 5; lvl <= 9; lvl++) { // minlevel = 3 (2.8pt/(0.5/ sqrt (2)))We initialize the grid.
N = 1 << (lvl);
init_grid (N);
run();
}
}Boundary conditions
Properties
event properties (i++)
{
foreach_face()
muv.x[] = fm.x[];
}Initial conditions
We set the viscosity field in the event properties.
mu = muv;The gravity vector is aligned with the channel.
const face vector g[] = {1., 1.};
a = g;We use “third-order” face flux interpolation.
#if ORDER2
for (scalar s in {u, p})
s.third = false;
#else
for (scalar s in {u, p})
s.third = true;
#endif // ORDER2
#if TREEWhen using TREE and in the presence of embedded boundaries, we also define the gradient of u at the cell center of cut-cells.
foreach_dimension()
u.x.embed_gradient = u_embed_gradient_x;
#endif // TREEWe initialize the embedded boundary.
#if TREEWhen using TREE, we refine the mesh around the embedded boundary.
astats ss;
int ic = 0;
do {
ic++;
p_shape (cs, fs);
ss = adapt_wavelet ({cs}, (double[]) {1.e-30},
maxlevel = (lvl), minlevel = (lvl) - 2);
} while ((ss.nf || ss.nc) && ic < 100);
#endif // TREE
p_shape (cs, fs);We also define the volume fraction at the previous timestep csm1=cs.
csm1 = cs;We define the no-slip boundary conditions for the velocity.
u.n[embed] = dirichlet (0);
u.t[embed] = dirichlet (0);
p[embed] = neumann (0);
uf.n[embed] = dirichlet (0);
uf.t[embed] = dirichlet (0);We finally initialize the reference velocity field.
foreach()
un[] = u.y[];
}Embedded boundaries
Outputs
We look for a stationary solution.
double du = change (u.y, un);
if (i > 0 && du < 1e-6)
return 1; /* stop */
}
event logfile (t = end)
{The total (e), partial cells (ep) and full cells (ef) errors fields and their norms are computed.
scalar e[], ep[], ef[];
foreach() {
if (cs[] == 0.)
ep[] = ef[] = e[] = nodata;
else {
double xy = (y - x) >= 0. ? (y - x) - 0.25 : (y - x) + 0.75;
xy /= sqrt (2.);
e[] = sqrt (sq (u.x[] - cos (pi/4.)*(exact (xy))) +
sq (u.y[] - sin (pi/4.)*(exact (xy))));
ep[] = cs[] < 1. ? e[] : nodata;
ef[] = cs[] >= 1. ? e[] : nodata;
}
}
norm n = normf (e), np = normf (ep), nf = normf (ef);
fprintf (stderr, "%d %.3g %.3g %.3g %.3g %.3g %.3g %d %g %g %d %d %d %d\n",
N,
n.avg, n.max,
np.avg, np.max,
nf.avg, nf.max,
i, t, dt,
mgp.i, mgp.nrelax, mgu.i, mgu.nrelax);
fflush (stderr);
if (lvl == 5) {
draw_vof ("cs", "fs", filled = -1, fc = {1,1,1});
cells ();
save ("mesh.png");
draw_vof ("cs", "fs", filled = -1, fc = {1,1,1});
squares ("u.x", spread = -1);
save ("ux.png");
draw_vof ("cs", "fs", filled = -1, fc = {1,1,1});
squares ("u.y", spread = -1);
save ("uy.png");
draw_vof ("cs", "fs", filled = -1, fc = {1,1,1});
squares ("p", spread = -1);
save ("p.png");
draw_vof ("cs", "fs", filled = -1, fc = {1,1,1});
squares ("e", spread = -1);
save ("e.png");
foreach() {
fprintf (stdout, "%g %g %g %g %g %g\n",
x, y,
u.x[], u.y[], p[],
e[]);
fflush (stdout);
}
}
}Results
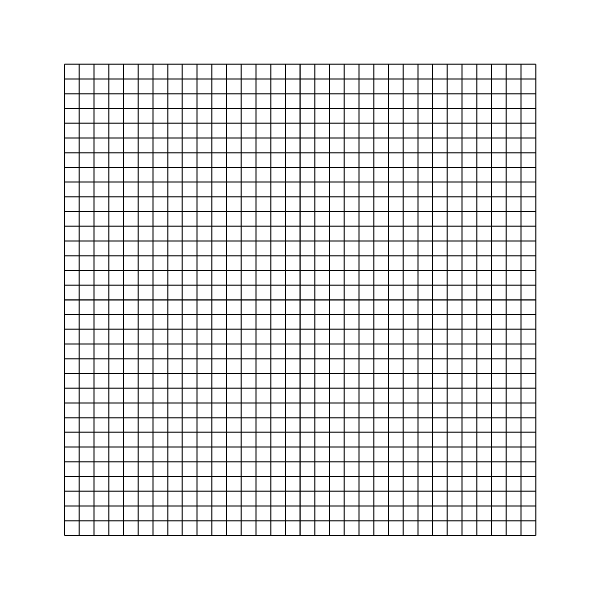
Mesh for l=5
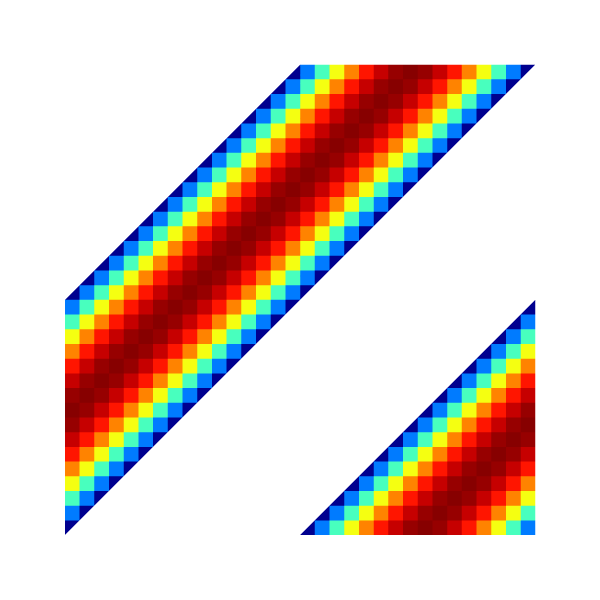
Velocity u.x for l=5

Velocity u.y for l=5
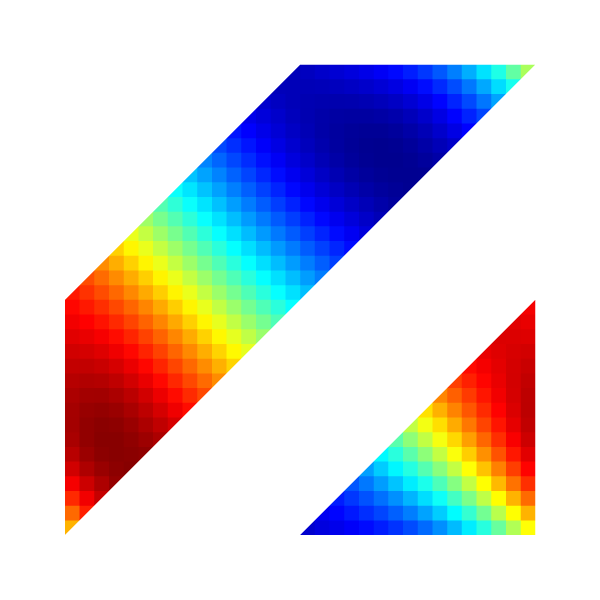
Pressure field for l=5

Error field for l=5
Errors
reset
set terminal svg font ",16"
set key bottom left spacing 1.1
set xtics 16,4,1024
set grid ytics
set ytics format "%.0e" 1.e-14,100,1.e2
set xlabel 'N'
set ylabel '||error||_{1}'
set xrange [16:1024]
set yrange [1.e-14:1.e-1]
set logscale
plot 'log' u 1:4 w p ps 1.25 pt 7 lc rgb "black" t 'cut-cells', \
'' u 1:6 w p ps 1.25 pt 5 lc rgb "blue" t 'full cells', \
'' u 1:2 w p ps 1.25 pt 2 lc rgb "red" t 'all cells'Average error convergence (script)
set ylabel '||error||_{inf}'
plot '' u 1:5 w p ps 1.25 pt 7 lc rgb "black" t 'cut-cells', \
'' u 1:7 w p ps 1.25 pt 5 lc rgb "blue" t 'full cells', \
'' u 1:3 w p ps 1.25 pt 2 lc rgb "red" t 'all cells'Maximum error convergence (script)
Order of convergence
We recover here the expected second-order convergence when using an adaptive mesh. On a uniform grid, the solution is almost exact.
reset
set terminal svg font ",16"
set key bottom left spacing 1.1
set xtics 16,4,1024
set ytics -4,2,4
set grid ytics
set xlabel 'N'
set ylabel 'Order of ||error||_{1}'
set xrange [16:1024]
set yrange [-4:4.5]
set logscale x
# Asymptotic order of convergence
ftitle(b) = sprintf(", asymptotic order n=%4.2f", -b);
Nmin = log(256);
f1(x) = a1 + b1*x; # cut-cells
f2(x) = a2 + b2*x; # full cells
f3(x) = a3 + b3*x; # all cells
fit [Nmin:*][*:*] f1(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($4)) via a1,b1; # cut-cells
fit [Nmin:*][*:*] f2(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($6)) via a2,b2; # full-cells
fit [Nmin:*][*:*] f3(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($2)) via a3,b3; # all cells
plot '< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:4 w lp ps 1.25 pt 7 lc rgb "black" t 'cut-cells'.ftitle(b1), \
'< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:6 w lp ps 1.25 pt 5 lc rgb "blue" t 'full cells'.ftitle(b2), \
'< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:2 w lp ps 1.25 pt 2 lc rgb "red" t 'all cells'.ftitle(b3)Order of convergence of the average error (script)
set ylabel 'Order of ||error||_{inf}'
# Asymptotic order of convergence
fit [Nmin:*][*:*] f1(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($5)) via a1,b1; # cut-cells
fit [Nmin:*][*:*] f2(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($7)) via a2,b2; # full-cells
fit [Nmin:*][*:*] f3(x) '< sort -k 1,1n log | awk "!/#/{print }"' u (log($1)):(log($3)) via a3,b3; # all cells
plot '< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:5 w lp ps 1.25 pt 7 lc rgb "black" t 'cut-cells'.ftitle(b1), \
'< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:7 w lp ps 1.25 pt 5 lc rgb "blue" t 'full cells'.ftitle(b2), \
'< sort -k 1,1n log | awk "!/#/{print }" | awk -f ../data/order.awk' u 1:3 w lp ps 1.25 pt 2 lc rgb "red" t 'all cells'.ftitle(b3)Order of convergence of the maximum error (script)
